This is the page for the project funded under NSF award #1754097:
The Role of Meiotic-Stage Non-Coding RNA in the Modulation of
Anther & Pollen Development in Grasses
Overview | Background | Broader Impacts | Research Team | Publications | Data & Software
Project Overview
Background of the Project
This project is a continuation of an earlier, successful project focused on maize pollen fertility and anther development (NSF award 1649424). The team of investigators is highly experienced in anther development, maize and rice genetics and genomics, RNA and chromatin, whole-genome analyses, and computational approaches for analyzing and displaying these data.
The project will build on resources, technologies, and genetic stocks that were developed or are underway from our previous NSF-funded work, and integrate new approaches in imaging, developmental and molecular analysis, genomics, and bioinformatics.
The fundamental questions to be addressed are:
- What are the spatial and temporal accumulation patterns of mRNA and proteins in anther lobe cell types? In particular, how do pre-meiotic and meiotic cells differ from the soma?
- Which transcription factors guide the transitions required for cell fate setting and differentiation of cell type-specific properties during anther development?
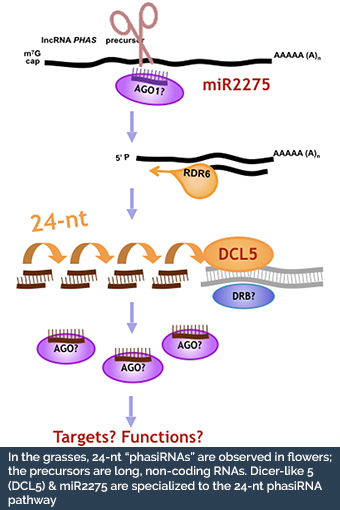
- What are the origins and functions of the non-coding phasiRNAs that are specifically expressed in grass inflorescences?

Mutant dcl1-5//Dcl1-5 anther development is temperature sensitive
Broader Impacts of Our Project
Together, maize and rice feed over half of humanity and are also the two species studied by these PIs. New information from this project will provide insights into the coordination of anther development and underlying cellular processes, including cellular remodeling, as plant cells switch from mitosis to meiosis.
New insights and resources of broad utility should ultimately contribute to controlling plant fertility in agriculture, particularly in the cereal crops.
The project builds and improves on the varied and extensive outreach and education components of the PIs and their contributions via scientific leadership to continue a strong, positive impact on local and scientific communities, and on the next generation of scientists. The laboratories working on this project offer numerous undergraduate- and graduate-student focused projects – please contact the co-PIs if you’re interested in working on the team.
Research Team
Publications
- Huang K, Demirci F, Meyers BC, Caplan JL. Quantitative, super-resolution localization of small RNAs with sRNA-PAINT. (2020) Nucleic Acids Research, in press. bioRxiv doi: 10.1101/716696v1.
- Bi H*, Fei Q*, Li R, Liu B, Xia R, Li T, Char SN, Meyers BC, Yang B. (2020) Disruption of miRNA sequences by TALENs induces varied length of miRNA production. Plant Biotechnology Journal, 18: 1526-1536. doi: 10.1111/pbi.13315 *equal contributions
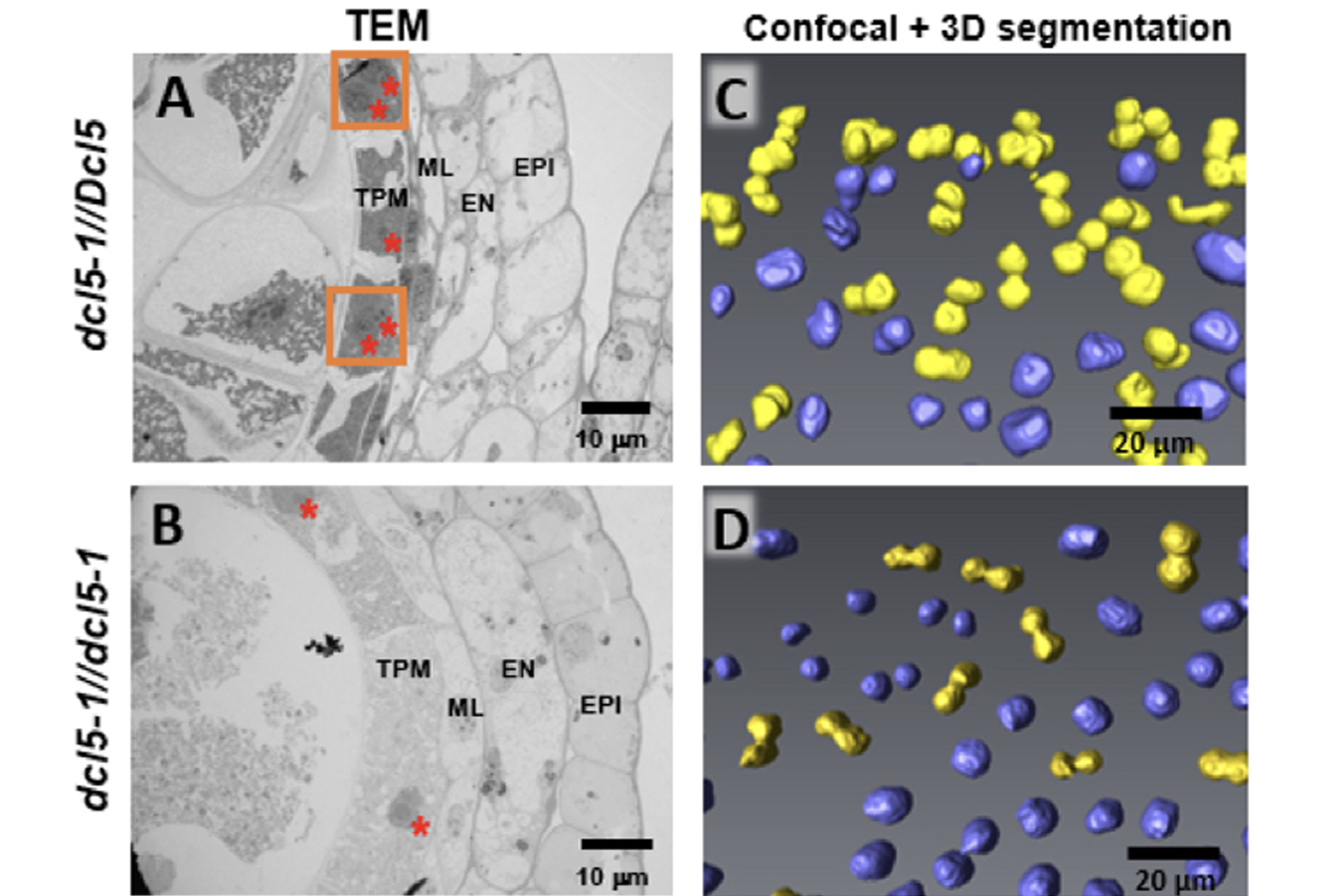
- Teng C*, Zhang H*, Hammond R, Kuang H, Meyers BC†, Walbot V†. (2020) Dicer-like 5 deficiency confers temperature-sensitive male sterility in maize. Nature Communications, 11: 2912. doi: 10.1038/s41467-020-16634-6 bioRxiv doi: 10.1101/498410 *equal contributions †co-corresponding authors
- Harkess A, Huang K, van der Hulst R, Tissen B, Caplan JL, Koppula A, Batish M, Meyers BC, Leebens-Mack J. (2020) Sex determination by two Y-linked genes in garden asparagus. The Plant Cell, 32: 1790-1796. doi: 10.1105/tpc.19.00859. bioRxiv doi: 10.1101/ 494112.
- Nakano M, McCormick K, Demirci C, Demirci F, Gurazada SGR, Ramachandruni D, Dusia A, Rothhaupt JA, Meyers BC. (2020) Next-gen sequence databases: RNA and genomic informatics resources for plants. Plant Physiology. 182: 136-146. doi: 10.1104/pp.19.00957
- Crisp PA, Hammond R, Zhou P, Vaillancourt B, Lipzen A, Daum C, Barry K, de Leon N, Buell RC, Kaeppler SM, Meyers BC, Hirsch CN, Springer NM. (2020) Variation and inheritance of small RNAs in maize inbreds and F1 hybrids. Plant Physiology. 182: 318-331. doi: 10.1104/pp.19.00817 bioRxiv doi: 10.1101/692400.
- van der Linde K, Walbot V. (2019) Pre-meiotic anther development. Curr Top Dev Biol. 131: 239-256. doi: 10.1016/bs.ctdb.2018.11.001
- Nelms B, Walbot V. (2019) Defining the developmental program leading to meiosis in maize. Science. 364: 52-56. doi:10.1126/science.aav6428
- Huang K, Baldrich P, Meyers BC†, Caplan J† (2019). sRNA-FISH: versatile
fluorescent in situ detection of small RNAs in plants. The Plant Journal, 98: 359-369. doi: 10.1111/tpj.14210 †co-corresponding authors. - Xia R†, Chen C, Pokhrel S, Ma W, Huang K, Patel P, Wang F, Liu Z, Li J, Meyers BC† (2019). 24-nt reproductive phasiRNAs are broadly present in angiosperms.Nature Communications 10: 627. doi: 10.1038/s41467-019-08543-0. †co-corresponding authors.
- Patel P, Mathioni S, Kakrana A, Shatkay H, Meyers BC (2018). Reproductive phasiRNAs in grasses are compositionally distinct from other classes of small RNAs. New Phytologist, 220: 851-864. doi: 10.1111/nph.15349. bioRxiv, doi: 10.1101/242727.
- Tamim S, Cai Z, Mathioni S, Zhai J, Teng C, Zhang Q, Meyers BC (2018). Cis-directed cleavage and nonstoichiometric abundances of 21-nt reproductive phasiRNAs in grasses. New Phytologist, 220: 865–877. doi: 10.1111/nph.15181. bioRxiv, doi: 10.1101/243907.
- Kakrana A., Mathioni SM, Huang K, Hammond R, Vandivier L, Patel P, Arikit S, Shevchenko O, Harkess AE, Kingham B, Gregory BD, Leebens-Mack JH, Meyers BC (2018). Plant 24-nt reproductive phasiRNAs from intramolecular duplex mRNAs in diverse monocots.Genome Research. 28: 1333-1344. doi: 10.1101/gr.228163.117.
- Bélanger S, Marchand S, Jacques P-E, Meyers BC, Belzile F (2018). Differential expression profiling of microspores during the early stages of isolated microspore culture using the responsive barley cultivar Gobernadora. G3, 8: 1603-1614. doi: 10.1534/g3.118.200208.
- Van Der Linde K, Timofejeva L, Egger R, Ilau B, Hammond R, Teng C, Meyers BC, Doehlemann G, Walbot V. (2018) Pathogen Trojan horse delivers bioactive host protein to alter maize (Zea mays) anther cell behavior in situ. The Plant Cell, 30: 528-542. doi: 10.1105/tpc.17.00238.
- Ma W, Chen C, Liu L, Zeng M, Meyers BC, Li, JG, Xia R. (2018)Coupling of microRNA-directed phasiRNA generation from long noncoding genes with alternative splicing and alternative polyadenylation in small RNA-mediated gene silencing. New Phytologist, 217:1535-1550. doi: 10.1111/nph.14934.
- Huang K, Doyle F, Wurz ZE, Tenenbaum SA, Hammond R, Caplan JL* & Meyers BC*. (2017) FASTmiR: An RNA-based sensor for in vitro and live-cell detection of small RNAs. Nucleic Acids Research. 45: e130 doi: 10.1093/nar/gkx504.
- Mathioni S, Kakrana A & Meyers BC. (2017) Characterization of plant small RNAs by next generation sequencing. Current Protocols in Plant Biology, 2:39-63. doi: 10.1002/cppb.20043.
- Fan Y, Yang J, Mathioni S, Yu J, Yang X, Wang L, Zhang Q, Shen J, Cai Z, Xu C, Li X, Xiao J, Meyers BC & Zhang Q (2016). PMS1T, producing phased small interfering RNAs, regulates photoperiod-sensitive male sterility in rice. Proc. Natl. Acad. Sci. USA (PNAS), 113(52):15144-49.
- Nan GL, Zhai J, Arikit S, Morrow D, Fernandez J, Mai L, Nguyen N, Meyers BC, & Walbot V (2016). MS23, a master basic helix-loop-helix factor, regulates the birth and maturation of tapetum. Development, 144(1):163-72.
- Fei Q, Yang L, Liang W, Zhang D* & Meyers BC (2016).Dynamic changes of small RNAs in rice spikelet development reveal specialized reproductive phasiRNA pathways. J. Exp. Botany, 67(21): 6037-6049. doi: 10.1093/jxb/erw361.
- Char SN, Neelakandan A, Nahampun H, Frame B, Main M, Spalding M, Becraft P, Meyers BC, Walbot V, Wang K, & Yang B (2016). An Agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize. Plant Biotechnology Journal, 15:257-268. doi: 10.1111/pbi.12611.
- Zhang Y, Xia R, Kuang H, & Meyers BC (2016). The diversification of plant NBS-LRR defense genes directs the evolution of microRNAs that target them. Mol Biol Evol, 33: 2692-2705. doi: 10.1093/molbev/msw154.
- Yang L, Qian X, Chen M, Fei Q, Meyers BC, Liang W, & Zhang D (2016). Regulatory role of a receptor-like kinase in specifying anther cell identity in rice. Plant Physiol, 171(3):2085-100. doi: 10.1104/pp.16.00016.
- Patel P, Ramachandruni D, Kakrana A, Nakano M & Meyers BC (2016). miTRATA: a web-based tool for microRNA truncation and tailing analysis.Bioinformatics. 32(3):450-2. doi: 10.1093/bioinformatics/btv583.
- Zhai J, Zhang H, Arikit S, Huang K, Nan GL, Walbot V & Meyers BC (2015). Spatiotemporally dynamic, cell-type-dependent premeiotic and meiotic phasiRNAs in maize anthers.Proc. Natl. Acad. Sci. USA (PNAS), 112(10): 3146-51. doi:10.1073/pnas.1418918112.
- Zhang H, Xia R, Meyers BC & Walbot V (2015). Evolution, functions and mysteries of plant ARGONAUTE proteins. Curr Opin Plant Biol, 27:84–90 doi:10.1016/j.pbi.2015.06.011.
- Thompson BE, Basham C, Hammond R, Ding Q, Kakrana A, Lee TF, Simon SA, Meeley R, Meyers BC & Hake S (2014). The dicerlike-1 homologue fuzzy tassel is required for the regulation of meristem determinacy in the inflorescence and vegetative growth in maize. Plant Cell, 26(12):4702-17. doi: 10.1105/tpc.114.132670.
- Kakrana A, Hammond R, Patel P, Nakano M & Meyers BC (2014). sPARTA: a parallelized pipeline for integrated analysis of plant miRNA and cleaved mRNA data sets, including new miRNA target-identification software. Nucleic Acids Res, 42(18):e139. doi:10.1093/nar/ gku693.
- Fei Q, Xia R & Meyers BC (2013). Phased, secondary siRNAs in post- transcriptional regulatory networks. Plant Cell, 25:2400–15.
- Arikit S, Zhai J & Meyers BC (2013). Biogenesis and function of rice small RNAs from non-coding RNA precursors.Curr Opin Plant Biol, 16:170-9.
- Patel P, Mathioni S, Kakrana A, Shatkay H, Meyers BC. Reproductive phasiRNAs in grasses are compositionally distinct from other classes of small RNAs. New Phytologist, 220: 851-864. doi: doi: 10.1111/nph.15349. bioRxiv, doi: 10.1101/242727.